| Jump to section |
Global Tea Genome Project | Funding | Dataset and framework | Web Sources Used |
| Reference | Principal Investigators | Visiting statistics | Contact us |
Global Tea Genome Project
For thousands of years, tea, this kind of oriental magical leaf, has been famous all over the world. The application and dissemination of tea is a model of the harmonious and sustainable development of man and plant. In the past two decades, the development of genomics technology has allowed us to explore the characteristics of plants and the course of their domestication from the perspective of heredity and genes. Yunnan is the region with the richest tea resources and is also considered the center of the origin of tea. Based on the protection, development, utilization of tea resources, and the responsibility for the sustainable development of tea, in the past ten years, our team has been committed to the research on the diversity and genome of tea genetic resources, and together with domestic and foreign counterparts, we have achieved a lot in tea genomes. However, the origin of global tea and the process of domestication of tea are still not fully understood. Here, we have completed the sequencing of thousands of tea genetic resources, especially the genome analysis of important tea resources around the world. These massive amounts of data are not only important for us to understand the origin and domestication of tea, but also for the breeding and protection of tea resources. This database not only includes the above-mentioned genomic information, but also information about tea and its related species. We hope that this database can become an important comprehensive database in the field of tea research.
Funding
Yunnan provincial key programs of Yunnan Eco-friendly Food International Cooperation Research Center project under grant 2019ZG00908
Yunnan Provincial Department of Science and Technology Biological Resources Digital Development and Application Project Number 202002AA100007
Dataset and framework
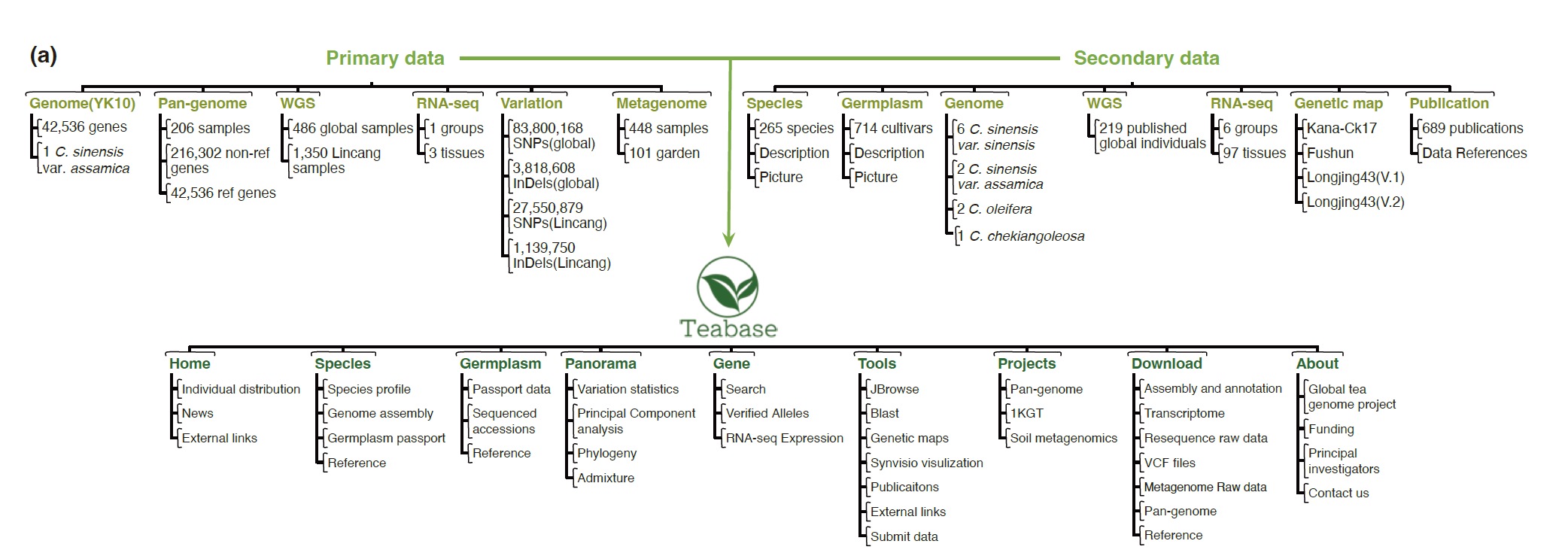
We built Teabase based on 12 Camellia genome assemblies, the resequencing data of 705 Camellia accessions from the global world, 100 RNA-seq datasets, and four genetic map datasets. The genome assembly of Camellia sinensis var. assamica contained in TeaBase is de novo assembled based on our data. High coverage re-sequencing data of 486 tea breeds worldwide are presented for the first time. In addition, Tea base provides three relatively complete projects, including the most comprehensive tea pan-genome to date, research on the genetic diversity of 1,350 tea trees in the central region of tea origin, and research on metagenomics of 448 soil of tea plants. These data and results are all reconstructed and analysed based on our data and are visualised and downloaded into this database for the first time. We also cover 265 species and 714 germplasm information of Camellia, even if they do not have enough omics data. We provide users and researchers with a complete data transmission strategy and invite all Camellia researchers around the world to improve Teabase. We collected a total of 2105 re-sequenced genomes for Camellia (up to Dec 31th, 2022). The newest Camellia genome assembly YK10(2020), is used as a reference. After quality control and data filtering, 486 globe genomes and 1350 lincang genomes are maintained in the population genomic analyses. The sequencing depth is >4X. And the genome coverage is >85%.
Web Sources Used
Echarts ( https://echarts.apache.org/zh/index.html )
d3 components ( https://d3js.org )
JavaScript ( https://www.javascript.com )
JBrowse ( http://jbrowse.org )
phylogeny.io ( https://github.com/oist/phylogeny-io )
JQuery ( https://jquery.com )
Apache web server ( http://www.apache.org )
ThinkPHP5.1 ( http://www.thinkphp.cn )
Codelgniter ( https://www.codeigniter.com/ )
Bootstrap ( https://getbootstrap.com )
MySQL( https://www.mysql.com )
Viroblast( https://indra.mullins.microbiol.washington.edu/viroblast/ )
SynVisio( https://synvisio.github.io/#/ )
Reference
Chen, J.-D., Zheng, C., Ma, J.-Q., Jiang, C.-K., Ercisli, S., Yao, M.-Z., and Chen, L. (2020) The chromosome-scale genome reveals the evolution and diversification after the recent tetraploidization event in tea plant. Hortic Res, 7, 63.
Gong, W., Xiao, S., Wang, L., Liao, Z., Chang, Y., Mo, W., et al. (2022) Chromosome-level genome of Camellia lanceoleosa provides a valuable resource for understanding genome evolution and self-incompatibility. The Plant Journal, 110, 881–898.
Lin, P., Wang, K., Wang, Y., Hu, Z., Yan, C., Huang, H., et al. (2022) The genome of oil-Camellia and population genomics analysis provide insights into seed oil domestication. Genome Biol, 23, 14.
Shen, T., Huang, B., Xu, M., Zhou, P., Ni, Z., Gong, C., et al. (2022) The reference genome of Camellia chekiangoleosa provides insights into Camellia evolution and tea oil biosynthesis. Horticulture Research, 9, uhab083.
Wang, P., Yu, J., Jin, S., Chen, S., Yue, C., Wang, W., et al. (2021) Genetic basis of high aroma and stress tolerance in the oolong tea cultivar genome. Horticulture Research, 8, 107.
Wang, Xinchao, Feng, H., Chang, Y., Ma, C., Wang, Liyuan, Hao, X., et al. (2020) Population sequencing enhances understanding of tea plant evolution. Nat Commun, 11, 4447.
Wang, F. et al. Chromosome-scale genome assembly of Camellia sinensis combined with multi-omics provides insights into its responses to infestation with green leafhoppers. Frontiers in Plant Science 13, (2022).
Wei, C., Yang, H., Wang, S., Zhao, J., Liu, C., Gao, L., et al. (2018) Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc Natl Acad Sci U S A, 115, E4151–E4158.
Xia, E., Li, F., Tong, W., Yang, H., Wang, S., Zhao, J., et al. (2019) The tea plant reference genome and improved gene annotation using long-read and paired-end sequencing data. Sci Data, 6, 122.
Xia, E., Tong, W., Hou, Y., An, Y., Chen, L., Wu, Q., et al. (2020) The Reference Genome of Tea Plant and Resequencing of 81 Diverse Accessions Provide Insights into Its Genome Evolution and Adaptation. Mol Plant, 13, 1013–1026.
Xia, E.-H., Zhang, H.-B., Sheng, J., Li, K., Zhang, Q.-J., Kim, C., et al. (2017) The Tea Tree Genome Provides Insights into Tea Flavor and Independent Evolution of Caffeine Biosynthesis. Mol Plant, 10, 866–877.
Zhang, Q.-J., Li, W., Li, K., Nan, H., Shi, C., Zhang, Y., et al. (2020) The Chromosome-Level Reference Genome of Tea Tree Unveils Recent Bursts of Non-autonomous LTR Retrotransposons in Driving Genome Size Evolution. Mol Plant, 13, 935–938.
Zhang, W., Zhang, Y., Qiu, H., Guo, Y., Wan, H., Zhang, X., et al. (2020) Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat Commun, 11, 3719.
Zhang, X., Chen, S., Shi, L., Gong, D., Zhang, S., Zhao, Q., et al. (2021) Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat Genet, 53, 1250–1259.
FOC
FOC (http://www.iplant.cn/)
Principal Investigators
 Jun Sheng
Jun Sheng
 Yang Dong
Yang Dong
 Wei Chen
Wei Chen
 Xuzhen Li
Xuzhen Li
Visiting statistics
Contact us
Institute: Yunnan Plateau Characteristic Agricultural Industry Research Institute
Address: No. 452, Fengyuan Road, Panlong District, Kunming, 650201, China
E-mail: dongyang@dongyang-lab.org