SMRT sequencing yields the chromosome-scale reference genome of tea tree, Camellia sinensis var. sinensis
Time:2020/1/2
Information: bioRxiv. 2020 Jan 2; doi: https://doi.org/10.1101/2020.01.02.892430
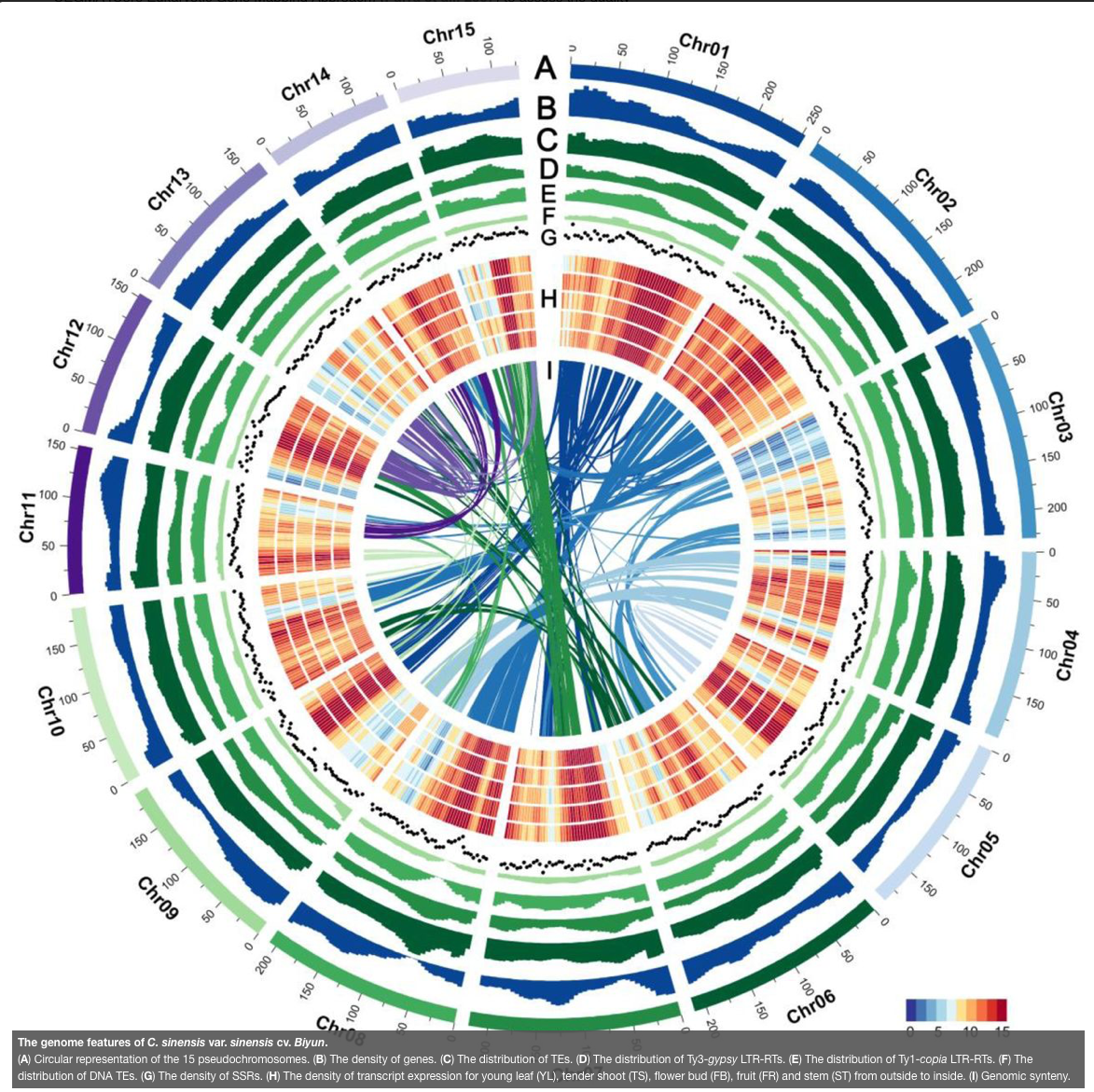
Tea is the oldest and most popular nonalcoholic beverage consumed in the world. It provides abundant secondary metabolites that account for its diverse flavors and health benefits. Here we present the first high-quality chromosome-length reference genome of C. sinensis var. sinensis using long read single-molecule real time (SMRT) sequencing and Hi-C technologies to anchor the ∼2.85-Gb genome assembly into 15 pseudo-chromosomes with a scaffold N50 length of ∼195.68 Mb. We annotated at least 2.17 Gb (∼74.13%) of repetitive sequences and high-confidence prediction of 40,812 protein-coding genes in the ∼2.92-Gb genome assembly. This accurately assembled genome allows us to comprehensively annotate functionally important gene families such as those involved in the biosynthesis of catechins, theanine and caffeine. The contiguous genome assembly provides the first view of the repetitive landscape allowing us to accurately characterize retrotransposon diversity. The large tea tree genome is dominated by a handful of Ty3-gypsy long terminal repeat (LTR) retrotransposon families that recently expanded to high copy numbers. We uncover the latest bursts of numerous non-autonomous LTR retrotransposons that may interfere with the propagation of autonomous retroelements. This reference genome sequence will largely facilitate the improvement of agronomically important traits relevant to the tea quality and production.